Structural Biology Lab
Studying relation
between structure of a biomolecule and its biological function!
Atta Ahmad (PhD)
N306 Howell Science Complex, ” We have a funded position for graduate
student for 2017-18
Department of Biology, Mail Stop 551 Please contact us for OPEN Positions”
East Carolina University,
Greenville, NC, 27858
Ph: 252-737-4777
Email: ahmada at ecu.edu
Research
My
lab is interested in studying protein structure, function and its relation to
human diseases. For some time DNA → RNA →
Proteins appeared to be the central dogma of life but due to Nobel prize
winning work of Christian Anfinsen (1972) and theoreticians like Cyrus
Levinthal (Levinthal’s paradox 1969) the importance of protein folding code
came to light and central dogma thus extended to DNA → RNA
→ Polypeptides → Functional Proteins. In other words,
the forces or code responsible for maintaining protein in its proper
3D-structure and thus efficient function gained impetus in the last part of
20th century. The profound influence of this concept would soon be evident as
mis-folded proteins (or toxic-folded) were found responsible for inclusion body
problems in biotechnology projects and mis-fold triggered ordered protein
aggregates “amyloid” were implicated in Prion, Alzheimer's and Parkinson's
diseases. The amyloid has since been found involved in more than 50 human
diseases. In addition, protein folding studies resulted in the discovery of a
group of enzymes and proteins called “chaperones” that are responsible for
helping maintain protein structure and function. We focus specifically on
following areas:
(if you don’t see the
picture please click here)
1) Protein structure and function: We have been
investigating the folding landscape of various proteins using chemical and thermal
agents. In the process we have identified new folding intermediates of proteins
and have contributed to the concept of mis-folding leading to amyloid formation
especially using insulin as a model protein. Our articles published in this
area have been cited more than 700 times.
2) Metal
toxicity: We have shown calcium induces amyloid type aggregation of peptides
Aβ40, Aβ42. Our studies further support the known implication of
calcium in Alzheimer's diseases through Aβ40 and Aβ42. Further, we
have shown copper induces structural modifications in protein α-synuclein
(implicated in Parkinson’s) again leading to its amyloid type aggregation. Both
metal ions exhibit this property at their physiological concentration and
therefore, these studies have immediate application in understanding the
important aspects that could further aid in the chelation therapy proposed for
Alzheimer’s and Parkinson's.
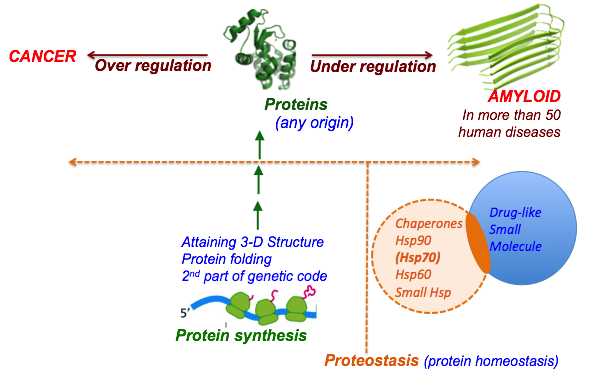
3) Hsp70 family Chaperone: Chaperones are groups of enzymes and proteins that help
other proteins attain structure essential for their function of which Hsp70 is
an important family. If a protein fails to fold Hsp70 tags it for degradation
(CHIP/ubiquitin pathway) and thus control the rate of
‘under-regulation/over-regulation’ of proteins or “proteostasis.”
It is not known how chaperones are hijacked into uncontrolled over production
of protein in the case of cancer or are not invoked during the mis-folded
accumulation of proteins inside the cells. Since these proteins have not been
investigated in detail the interactions between themselves and with substrate
proteins is also not well characterized. Our efforts, in addition to addressing
to each of these, also extends to screening chemical libraries to
discover efficient modulators of chaperone systems for their immediate
application as therapeutics to human disease systems mentioned
above.
Techniques: Biochemical, Biophysical, Molecular biology, (refer
to our publication for details)
|
New
Positions: “We have a funded
position for graduate student 2017-18” We are always looking for interested
graduate and undergraduate students, please feel free to contact us. Current Members Sidney Bedsole Undergraduate, ECU Ellen Styons Undergraduate, ECU |
Teaching BIOL
1050 - Biology for
non-majors BIOL
1100 BIOL
1150/51 BIOL
3030 - Principles
of physiology BIOL
5870 - Molecular
Biology of the Gene BIOL
6030/4650 - Biology of Cancer BIOL
4891/6244 - Biochemistry Lab BIOL
6504/6514 - Research Problems in
Biology BIOL
7000 - Dissertation/thesis |
|
Previous members Student Kong Peng Lor Master’s Student Hamza Karimi Research Associate Christopher Lovick Undergraduate Student David F Jahad Master’s Student Caleb
Stratton Master’s
Student Paola Perez-May Undergraduate
student Aston C Fuller Undergraduate student JoColl Burgess Undergraduate
student |
Publications [pubmed list]
(* Indicates corresponding author; #ECU
students; $ECU faculty)
At ECU:
21. Ahmad A*, Jahad DF#, Karimi H# (2020) An
economical protocol for gene transformation. (communicated)
22.
Akbarian M, Kianpour M, Ahmad A*,
Yousefi R*, Ali Akbar Moosavi-Movahedi.
(2020) Convergence of
conformational populations of insulin under two scenarios: the urea-induced and
seeds-mediated fibrillations. (communicated)
23. Akbarian M, Yousefi R, Mossavi-Movahedi A, Ahmad
A, Uversky VN. (2019) Modulating insulin fibrillation using engineered B-chains
with mutated C-termini: new insight to insulin fibrillation. Biophysical Journal 117, 1626-41
24. Ahmad A*, Jahad, DF#, Karimi
H#, Farwell
MA$, Scemama J$ and
Putnam-Evans C$. (2019). Modifications
in the Transformation Step of Commercially Available Site-directed Mutagenesis
Kit Increase Its Price-performance. Bio-101:
e3315.
25. Taylor IR, Ahmad A, Wu T, Nordhues
BA, Bhullar A, Gestwicki JE, Zuiderweg
ERP. (2018) The disorderly conduct of Hsc70 and its interaction with the
Alzheimer’s related Tau protein J Biol
Chem. 293,10796
26. Morozova
K, Clement CC, Kaushik S, Stiller B, Arias E, Ahmad A, Rauch JN,
Chatterjee V, Melis C, Scharf B, Gestwicki
JE, Cuervo AM, Zuiderweg ER, Santambrogio
L. (2016) Hsc-70 Structural and
Biological Interaction with Phosphatidylserine in Endosomal Microautophagy. J Biol Chem. 291,
18096-106
27. Ahmad
A*, Muzaffar M (2016) Molecular Chaperones and Co-chaperones as Therapeutic Targets
for Cancer. J Mol Pharm Org Process
Res. 4:e124
[doi: 10.4172/2329-9053.1000e124]
28. Ahmad A*, Stratton CM#, Scemama JL$, Muzaffar M$. (2016) Effect of Ca2+
on Aß40 fibrillation is characteristically different. Int J Biol Macromol.
89, 297-304
29. Ahmad A*, Zuiderweg ERP DnaK/DnaJ complex in ATP state (in preparation)
30. Ahmad A* Understanding
the process of Aβ40/Aβ42 aggregation (in preparation)
31. Ahmad A*
Biophysical characterization of structural contribution of BSA towards its
amyloid (in preparation)
Before ECU
1. Li X, Srinivasan S, Connarn
J, Ahmad A, Young Z, Kabza A, Zuiderweg E, Sun D, Gestwicki JE. (2013) Analogs of the Allosteric Heat Shock
Protein 70 (Hsp70) Inhibitor MKT-077 as Anti-Cancer Agents. ACS Med Chem Lett 4, 1042-47
2. Cesa LC, Patury S, Komiyama T, Ahmad A, Zuiderweg ER and Gestwicki JE. (2013) Inhibitors of Difficult
Protein-Protein Interactions Identified by High-Throughput Screening of
Multiprotein Complexes. ACS Chem 8,
1988-97
3. Zuiderweg ERP,
Bertelsen EB, Rousaki A,
Mayer MP, Gestwicki JE and Ahmad A (2013) Allostery in the Hsp70 chaperone proteins. Top Curr Chem 328, 99-153
4. Ahmad A*, Burns CS, Fink AL and Uversky
VN (2012) Peculiarities of copper binding to a-synuclein. Journal of Biomol Struct Dyn 29,
825-842
5. Zuiderweg ERP
and Ahmad A (2012)
Evaluation of competing J domain: Hsp70 Complex Models in light of methods
used. Proc Natl Acad Sci USA 109, E735
6. Ahmad A, Bhattacharya A, McDonald RA, Cordes M, Ellington B,
Bertelsen EB and Zuiderweg
ERP (2011) The Hsp70 chaperone/DnaJ co-chaperone
complex employs an unusual dynamical interface. Proc Natl Acad Sci USA 108, 18966-18971
7. Muzaffar M and Ahmad A* (2011) The Mechanism of Enhanced
Insulin Amyloid Fibril Formation by NaCl Is Better Explained by a
Conformational Change Model. PLoS ONE 6(11): e27906
8. Ahmad A* (2010) DnaK/DnaJ/GrpE of Hsp70 system have
differing effects on alpha-synuclein fibrillation involved in Parkinson's
disease. Int J Biol Macromol. 46,
275-279.
9. Ahmad A*, Muzaffar M and Ingram VM (2009) Ca2+,
within the physiological concentrations, selectively accelerates Aβ42
fibril formation and not Aβ40 in vitro. Biochim. Biophys.
Acta. 1794,
1536-1547.
10. Munishkina
LA, Ahmad A, Fink AL and
Uversky VN (2008) Guiding protein aggregation by
molecular crowding. Biochemistry 47, 8993-9006.
11. Hood EE, Love R, Lane J, Bray J, Clough
R, Pappu K, Drees C, Hood KR, Yoon S, Ahmad A, Howard JA (2007)
Subcellular targeting is a key condition for high-level accumulation of
cellulase protein in transgenic maize seed. Plant Biotechnol J. 5, 709-19.
12. Hong DP, Ahmad A and Fink AL (2006) Fibrillation of human insulin A
and B chain. Biochemistry 45, 9342-9353.
13. Ahmad A, Uversky VN, Hong D and
Fink AL (2005) Early events in insulin monomer fibrillation J Biol. Chem. 280,
42669-42675.
14. Ahmad A, Millet IS, Doniach S, Uversky VN and Fink AL (2004) Stimulation of insulin
fibrillation by urea-induced intermediates. J Biol.
Chem. 279,
14999-13.
15. Ahmad A, Millet IS, Doniach S, Uversky VN and Fink AL (2003) Partially Folded
Intermediates in Insulin Fibrillation. Biochemistry 42, 11404-11416.
16. Akhtar MS, Ahmad A, and Bhakuni V (2002)
Guanidinium chloride and urea induced unfolding of dimeric enzyme glucose
oxidase. Biochemistry 41, 3819-3827.
17. Akhtar MS, Ahmad A and Bhakuni V (2002)
Divalent cation induced changes in structural properties of the dimeric enzyme
glucose oxidase: Dual effect of dimer dissociation with loss of cooperative
interactions in enzyme monomer. Biochemistry
41, 7142-7149.
18. Ahmad A, Akhtar MS and Bhakuni V (2001)
Monovalent Cation Induced Conformational Change in Glucose Oxidase Leading to
Stabilization of Enzyme. Biochemistry 40, 1945-1955.
19. Ahmad A, Madhusudanan KP and Bhakuni V (2000) Trichloroacetic Acid and Trifluoroacetic
Acid Induced Unfolding of Cytochrome c: Stabilization of a Native like Folded
Intermediate. Biochim. Biophys. Acta. 1480, 201-210.
20. Ali V, Prakash K, Kulkarni S, Ahmad A, Madhusudanan
KP and Bhakuni V (1999)
8-Anilino-1-Napthalenesulfonic Acid Induces Folding of Acid Unfolded Cytochrome
c to Molten Globule State as a Result of Electrostatic Interactions. Biochemistry 38,
13635-13642.
Funding
Role: PI
Grant/project –$60,000 $45,000
Title: "Proposal
for upgrading Biochemistry lab, HPLC system."
Agency: ITCS
(Information Technology and computing services) ECU –
Status: Awarded – 07/07/2018
Role: PI
Grant/project –$25,000
Title: "Proposal
for upgrading Biochemistry lab, Multi-well plate reader."
Agency: ITCS
(Information Technology and computing services) ECU
Status: Awarded – 01/12/2018
Role: PI
Grant/project – F033032, $45,000
Title: "Examining role of
chaperones in protein quality control and its relation with diabetes."
Agency: MDRC Michigan - Status:
Awarded – 2/10/2013-11/30/2013
Role: PI
Grant/project – $45,000
Title: "Utilizing Chaperones
for intervention in cancer as therapeutic and biomarker targets."
Agency: Golfers Against Cancer,
Greensboro, NC Chapter - Status: Awarded 5/1/2015 - 4/30/2016
Role: Co-PI
Grant/project - $5000
Title: “Exploring PD-1, PD-L1
interactions with Hsp70 for mechanistic insight and drug target identification
in triple negative breast cancer”
Agency: ECU graduate research program
– Status: Awarded 8/7/2015 – 7/1/2016
Funding
Links